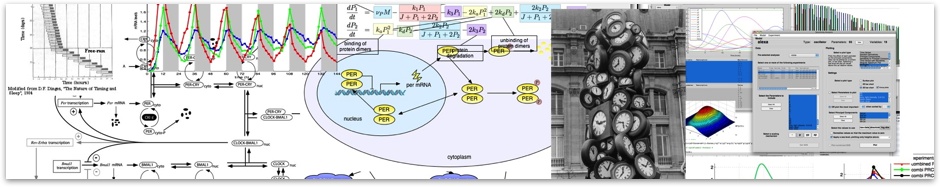
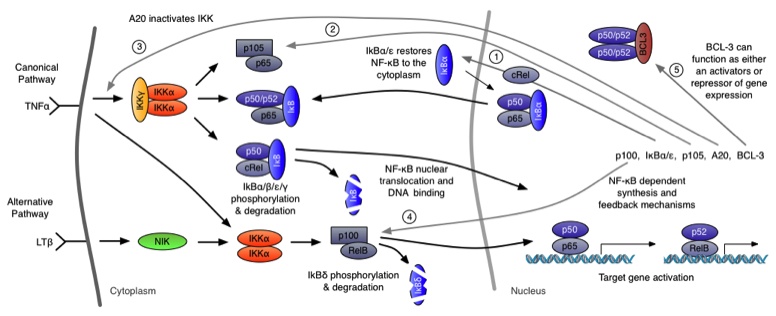
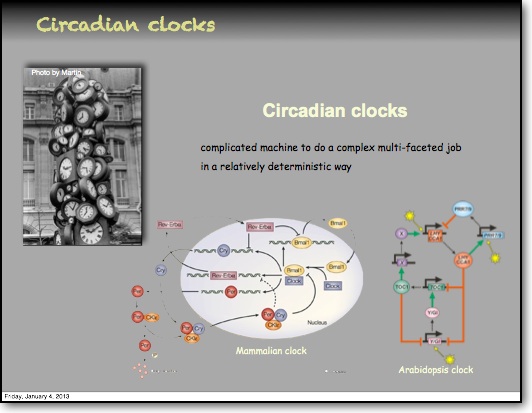
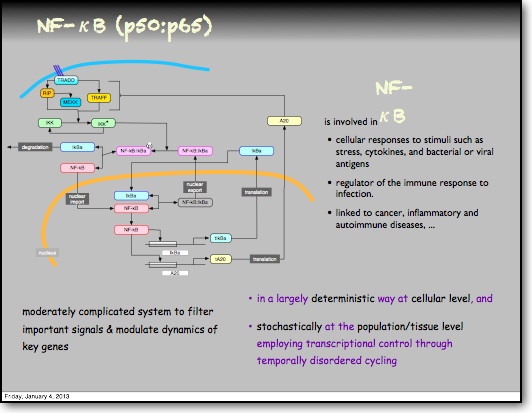
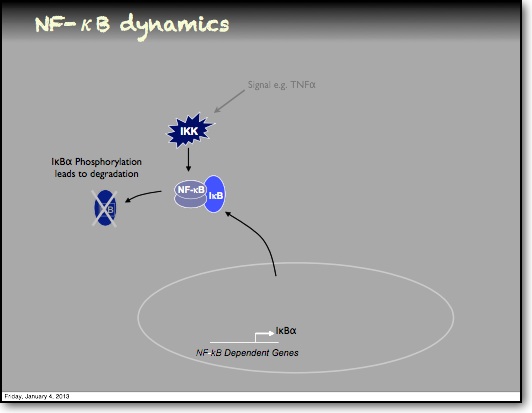
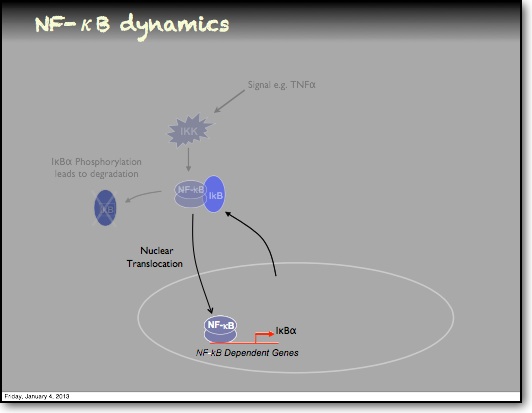
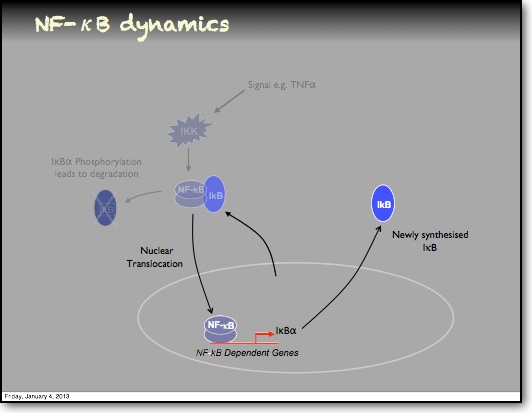
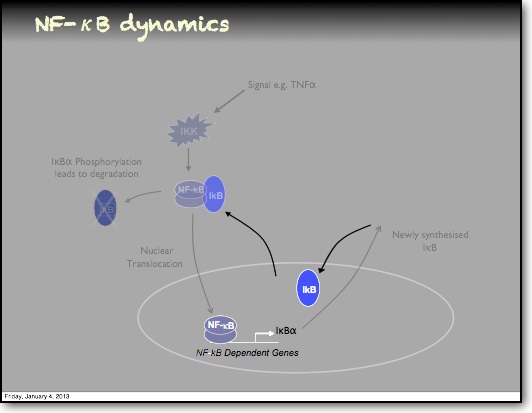
In the RelA-p50 dimer, the p50 protein does not contain a transcriptional activation domain, but p50 homodimers are associated with gene repression. p50 is produced following processing of p105. It has a N-terminal IκB domain (IκBγ), which is degraded soon after p105 processing. Normally this is believed to be a constitutive process, but it may also be a response to cytokine stimulation. One enzyme that has been implicated in the control of p105 processing is GSK3β. This enzyme phosphorylates and stabilises p105 (preventing constitutive processing) in the absence of other stimuli and primes p105 for IKK-induced degradation. The activation of the RelA-p50 dimer is called the canonical pathway.
Feedback and the timing of oscillations
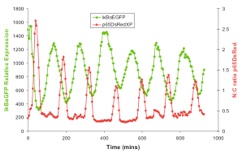
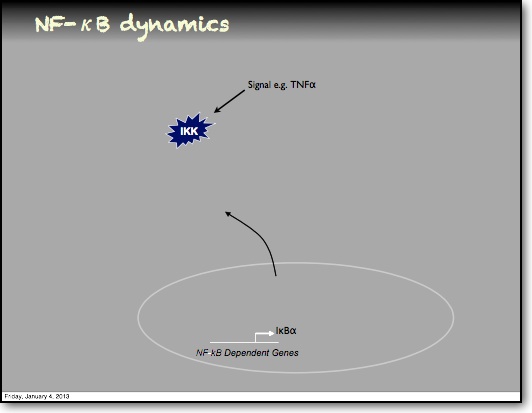
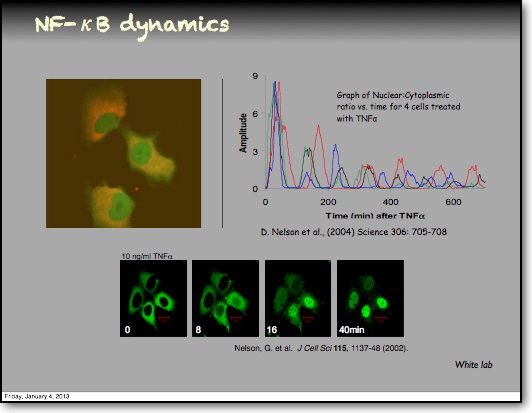
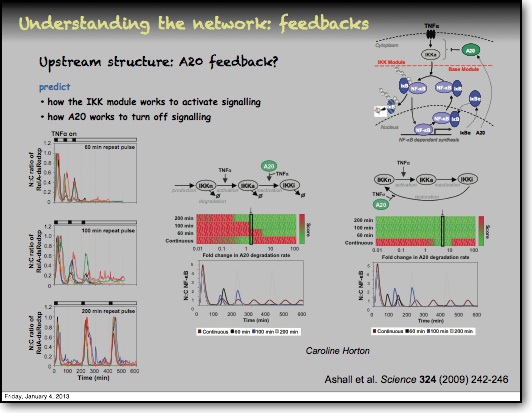
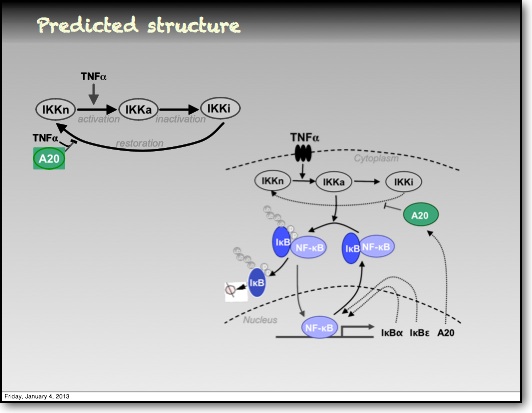
In addition to the IκBα and IκBε there are other feedbacks from a set of genes for proteins that act upstream from IKK, the most studied being the protein A20. The timing of oscillations in response to TNFα is remarkably consistent between cells. However, slight variation in the timing between peaks of translocation causes cells to quickly fall out of synchronisation with each other2. It has been proposed that transcriptional noise in negative feedback genes (there being only 2 gene copies of each) may explain this lack of synchronicity between the cells. We used a combined modelling and experimental strategy to suggest that the transcriptional activation of the IκBε feedback loop is delayed relative to the IκBα feedback loop by around 40 min. Analysis of the mathematical model suggested that this timing lay at a theoretical optimum for the generation of heterogeneity between cells and thus might be an evolutionarily selected phenotype. Such heterogeneity might minimise the overall fluctuations in cytokine levels in a tissue and therefore be important in accurate control of the tissue-level inflammation. This raises the interesting idea that overall tissue level inflammation may be in part regulated by the degree of heterogeneity between individual cells. This potentially represents a new phenotype that might be a possible drug target to manipulate the balance between hypo- and hyper-inflammation.