David Rand
Warwick Systems Biology & Mathematics Institute

Circadian clocks

Linnaeus' flower clock was a garden plan hypothesized by Carolus Linnaeus that would take advantage of several plants that open or close their flowers at particular times of the day to accurately indicate the time.
My research in this area is very much concerned with understanding the design principles of circadian clocks. What are their roles and how is this reflected in their structure? I would add that since the clock is a dynamical system with a very complex molecular structure and since the role of the clock is clearly multifacited, this cannot be answered without the use of mathematics. My collaborators are listed here.
The circadian clock adapts organisms to the environmental day-night cycle both behaviorally and physiologically. In animals, not only are complex behaviors such as sleep and mood governed by this oscillator, but also different body functions such as digestion, circulation, and respiration. The basic mechanism of this clock is cell-autonomous in all studied species possessing a circadian clock i.e. each cell contains a network of genes and proteins that can generate an oscillator with a period of approximately 24 hours. These oscillators can, in turn be entrained, to the relevant 24-hour environmental day-night cycle.
Mammalian timing system
In mammals, individual clocks in most cells are synchronized by a brain “master clock” in the suprachiasmatic nucleus of the hypothalamus. This orchestrates all rhythmic physiology and signals from this master clock entrain the clocks in peripheral cells. This global system is often referred to a the Circadian Timing System. It should be thought of as the body'd metronome as explained in this INSERM article.
Human diseases
In humans, disruption of the circadian rhythm is associated with sleep disorders and many other diseases including cancer, obesity and diabetes. On a cellular level, circadian physiology extends even to processes such as proliferation, apoptosis, and DNA damage repair, which are thought to play important roles in cancer initiation and control.
Plants and other species
In plants there is no such "master clock" but the cellular circadian clock is entrained by the cycles of light and dark, and temperature. The light input pathway to the clock is complex and only just becoming understood.
Besides vertebrates and plants most many other species have such circadian clocks including flies, fungi, and bacteria.
In individual cells, the circadian clock mechanism consists of oscillating feedback loops of transcription of “core” oscillator genes and posttranslational modifications of their protein products that regulate protein stability, activity, and/or localization.
Mammalian clock network
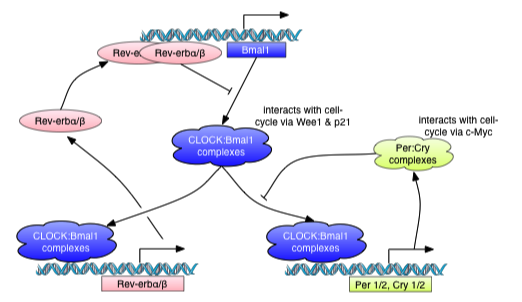
The majority of clock components are transcriptional activators or repressors that modulate protein stability and nuclear translocation, and create two interlocking feedback loops. In the first, CLOCK and BMAL1, heterodimerize in the cytoplasm to form a complex that, following translocation to the nucleus, initiates transcription of target genes such as the core clock genes Period genes (PER1, PER2, and PER3) and two Cryptochrome genes (CRY1 and CRY2).
Negative feedback is achieved by PER:CRY heterodimers that translocate back to the nucleus to repress their own transcription by inhibiting the activity of the CLOCK:BMAL1 complexes. Another regulatory loop is induced when CLOCK:BMAL1 heterodimers activate the transcription of Rev-ErbA and Rora, two retinoic acid-related orphan nuclear receptors. REV-ERBa and RORa subsequently compete to bind retinoic acid-related orphan receptor response elements (ROREs) present in Bmal1 promoter. Through the subsequent binding of ROREs, members of ROR and REV-ERB are able to regulate Bmal1. While RORs activate transcription of Bmal1, REV-ERBs repress the same transcription process. Hence, the circadian oscillation of Bmal1 is both positively and negatively regulated by RORs and REV-ERBs.
Plant clock network
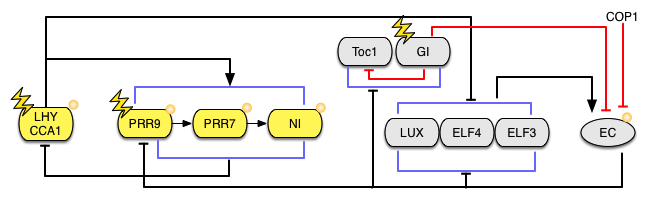
The two morning-specific MYB transcription factors, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED-1 (CCA1) act to repress expression of a pseudo response regulator (PRR1, also known as TIMING OF CAB-1, or TOC1) during the day, by binding to an Evening Element (EE) motif in the promoter of its gene.
As LHY/CCA1 protein levels decline towards the evening, TOC1 accumulates and acts to repress transcription from their respective promoters. Expression of LHY and CCA1 mRNAs resumes towards dawn, when this repression is lifted, through down-regulation of TOC1 transcription by an Evening Complex (EC) composed of three proteins, LUX and EARLY FLOWERING (ELF) 3 and 4. In addition to TOC1, three other PRR proteins (PRR9, 7 and 5) are expressed sequentially throughout the day (4). Each of these proteins binds to the LHY and CCA1 promoters to repress their activity, and their combined effect over the course of the circadian cycle ensures that this inhibition is maintained from the late morning until the following dawn (5).
Why are clocks so complicated and not just simple oscillators?
A key problem that I am trying to address is why the regulatory networks of these clocks almost universally have a highly complicated structure, with multiple interlocking positive and negative feedback loops when a simple negative feedback loop can reliably produce robust oscillations. My basic hypothesis is that it is to enable the flexibility to respond in multiple ways to multiple evolutionary demands some of which will conflict such as in the following examples
Ability to regulate a very large number of target genes
The clock regulates a very large number of genes - typically, in a given tissue the clock regulates on the order of 10-20% of the genes.
Flexibility to respond in multiple ways to different environments and tissues
The clock needs to adjust its behaviour to appropriately control its target genes in ways that will depend upon the tissue that the cell is in and the environment that the organism is in.
Robust entrainment
Its structure needs ti enable robust entrainment by environmental signals such as light and temperature with correct phases for multiple output pathways.
Temperature compensation
It is very important that it should be temperature compensated i.e. the behaviour of the clock should be buffered against changes in the overall temperature of the environment. It also needs to be buffered against other environmental variations such as those of pH, nutrition or growth conditions.
Photoperiodism
It must must give appropriate responses for the different environmental conditions arising during the course of a year
Reliability and robustness
It should be robust to extrinsic cellular noise so that period and phase relationships should not fluctuate too much. It should be resistant to noise in the sense that it function reliably with appropriate (small) numbers of molecules.
It is difficult, perhaps impossible, to undestand the design principals without mathematics
Such an approach is developed in the following papers
A new mathematical approach to uncovering the link berween structure and function for circadian clocks
Design principles underlying circadian clocks.
D. A. Rand , B. V. Shulgin, D. Salazar , A. J. Millar, Journal of The Royal Society, Interface 1 (2004) online at DOI: 10.1098/rsif.2004.0014
A new mathematical approach to the design principles of circadian clocks
Uncovering the design principles of circadian clocks: Mathematical analysis of flexibility and evolutionary goals.
D.A. Rand, B.V. Shulgin, J.D. Salazar and A.J. Millar Journal of Theoretical Biology, 238(3) (2006) 616-635.
Mathematical analysis of temperature compensation for the Neurospora crassa circadian clock that integrates a large amount of data.
Isoform switching facilitates period control in the Neurospora crassa circadian clock
O. E. Akman, J.C.W. Locke, S. Tang, I. Carré, A. J. Millar & D. A. Rand, Molecular Systems Biology. 4:164 http://www.nature.com/doifinder/10.1038/msb.2008.5 doi:10.1038/msb.2008.5
A new approach to sensitivity analysis
Mapping the global sensitivity of cellular network dynamics: Sensitivity heat maps and a global summation law.
D. A. Rand. J. R. Soc. Interface (2008) 5 S59-S69 doi:10.1098/rsif.2008.0084.focus
New summation theorems that substantially generalise previous results to dynamic non-stationary solutions such as periodic orbits and transient signals and apply to both autonomous and non-autonomous systems such as forced nonlinear oscillators.
Network control analysis for time-dependent dynamical states.
D. A. Rand. Dynamics and Games in Science, in honour of Mauricio Peixoto and David Rand. Springer 2010.
Modelling the photoperiod switch in plants predicts new role for FKF1.
Prediction of Photoperiodic Regulators from Quantitative Gene Circuit Models.
J. D. Salazar, T. Saithong, P. E. Brown, J. Foreman, J. C. W. Locke, K. J. Halliday, I. A. Carre, D. A. Rand and A. J. Millar. Cell 139, 1170–1179, DOI 10.1016/j.cell.2009.11.029
Analysis of a new model for the Neurospora circadian clock
Robustness from flexibility in the fungal circadian clock.
O. E. Akman, D. A. Rand, P. E. Brown and A. J. Millar. BMC Systems Biology 2010, 4:88
Clocks need to track more than one phase
Quantitative analysis of regulatory flexibility under changing environmental conditions.
K. D. Edwards, , O. E. Akman, K. Knox, P. J. Lumsden, A. W. Thomson, P. E. Brown, A. Pokhilko, L. Kozma-Bognar, F. Nagy, D. A. Rand, and A. J. Millar, Molecular Systems Biology 6:424.
The basic mathematical tools you need for experimental design and sensitivity analysis for stochastic regulatory or signalling systems. Uses the linear noise approximation.
Sensitivity of stochastic chemical kinetics models.
M. Komorowski, M. Costa, D. A. Rand, and M. L. Stumpf, PNAS 2011 108 (21) 8645-86
Using new data and mathematical modelling and analysis we test two hypotheses: that the targets of light regulation are sufficient to mediate temperature compensation and that, rather than using specific molecular mechanisms to achieve temperature compensation, the plant clock uses non-specific network balancing.
Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures.
Peter D Gould, Nicolas Ugarte, Mirela Domijan, Maria Costa, Julia Foreman, Dana MacGregor, Ken Rose, Jayne Griffiths, Andrew J Millar, Bärbel Finkenstädt, Steven Penfield, David A Rand, Karen J Halliday & Anthony J W Hall. Molecular Systems Biology 9 Article number: 650 doi:10.1038/msb.2013.7
State of the art algorithms to analyse circadian data.
Inference on periodicity of circadian time series.
Maria J. Costa, Bärbel Finkenstädt, Veronique Roche, Francis Levi, Peter D. Gould, Julia Foreman, Karen Halliday, Anthony Hall, David. A. Rand. Biostatistics (2013) 14 (4): 792-806 first published online June 6, 2013 doi:10.1093/biostatistics/kxt020
We dynamically measured the temperature coefficient, Q10, of mRNA synthesis and degradation rates of the Arabidopsis transcriptome. Our data show that less frequent chromatin states can produce temperature responses simply by virtue of their rarity and the difference between their thermal properties and those of the most common states.
Direct measurement of transcription rates reveals multiple mechanisms for configuration of the Arabidopsis ambient temperature response.
Kate Sidaway-Lee, Maria J. Costa, David Rand, Bärbel Finkenstadt, and Steven Penfield. Genome Biology 2014, 15:R45 (3 March 2014)
In the absence of other signals, the cell cycle and circadian clock robustly phase-lock each other in a 1:1 fashion so that in an expanding cell population the two oscillators oscillate in a synchronised way with a common frequency. However, there are additional clock states: as well as the low-period phase-locked state there are distinct coexisting states with a significantly higher period clock and a different frequency ratio.
Phase-locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle.
C. Feillet, P. Krusche, F. Tamanini, R. C. Janssens, M. J. Downey, P. Martin1, M. Teboul1, S. Saito, F. Levi, T. Bretschneider, G. T. J. van der Horst, F. Delaunay, D. A. Rand. PNAS (to appear)